The provided services are based on tools that we previously developed and shared via GitHub. The purpose of this web server is to enable biologists without bioinformatics skills to benefit from these tools. All services are provided free of charge, but without any warranty. Many thanks to de.NBI for excellent support! While we aim to maintain this web server for the foreseeable future, some services may be discontinued without prior notice. Please cite the corresponding publications when using this web server for your studies. This is the best way to support this service. When you upload a FASTA file and click "submit", a job is scheduled for execution on our server. Once the necessary computational resources are available, the job will be executed. Most jobs should be completed within minutes and our server is capeable of running multiple jobs in parallel. Please avoid the submission of unnecessary jobs to protect the environment by reducing energy consumption.
Submitted FASTA files will be stored for the analysis. By submitting your job, you agree to this. Files associated with a completed job are stored for 48 hours to enable the download. Due to limited disk space, we cannot keep your files indefinitely. Please make sure to download your results as soon as possible.
There is an option to accept cookies which allows your browser to keep track of your submitted jobs. This option is recommended if you do not provide an email address. Once the job is completed, a download button will be available to retrieve a compressed archive with all result files.
There is another option to receive a download link via email once your job is completed. If you select this option, you get one email upon successfull submission of your job and one email upon completion. The link in the first email also allows you to check the progress of your job.
Feel free to contact us via email if you have any questions or comments related to our tools.
KIPEs (Knowledge-based Identification of Pathway Enzymes) is now available as a web service. This enables the automatic identification and annotation of flavonoid biosynthesis genes/proteins. User only need to supply a FASTA file with the peptide or transcript sequences. Please read the original publication for details Pucker et al., 2020. KIPEs is available for download from a GitHub repository.
Our automatic MYB annotation workflow is now available as a web service. Users can supply a FASTA file of peptide or transcript sequences to identify the MYB transcription factors. Please have a look at the original publication for details: Pucker, 2022. MYB annotator is available for download from a GitHub repository.
CoExp enables user to identify co-expressed genes. A list of genes can be provided and the analysis is performed for each of them. All genes with a sufficient co-expression with the candidate gene are returned. A functional annotation file can be provided to assign a functional annotation to the results. CoExp is available for download from a GitHub repository.
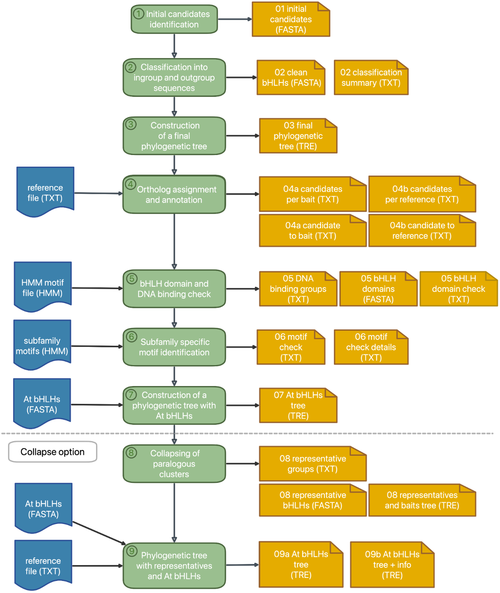
Our automatic bHLH annotation workflow is now available as a web service. Users can supply a FASTA file of peptide or transcript sequences to identify the bHLH transcription factors. Please have a look at the original publication for details: Thoben & Pucker, 2023. bHLH annotator is available for download from a GitHub repository.
NAVIP (Neighborhood-Aware Variant Impact Predictor) processes a variant calling format (VCF) file and predicts the functional impact of the sequence variants on annotated genes. NAVIP is available on our web server. Additional details can be found in the NAVIP GitHub repository.
Are you missing a feature or one of our tools? Please get in touch via email. We are planning to extend the collection of offered tools. Learning about the interest in different tools helps us to prioritize the addition of new tools.